Research
Segmental Domains
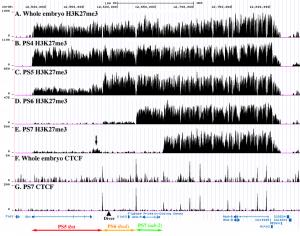
The Polycomb regulation of the bithorax complex differs from segment to segment, based on genetic studies by E. B. Lewis, and on our own work with enhancer traps and accessibility probes within the complex. Thus, molecular assays using cells from the whole organism show only profiles averaged across all segments. We have used combinations of genomic transgenes expressing the GAL4 activator and the GAL80 repressor of GAL4 to drive expression of a fluorescent marker in single segments (more exactly, in single “parasegments”). The Gal4 turns on a fluorescent version of a nuclear membrane protein, and fixed, fluorescent nuclei are collected on a FACS machine. The nuclei are then used for ChIP-seq analysis (shearing, immunoprecipitation, library construction, and deep sequencing); this part of the effort has been carried out by Sarah Bowman, Aimee Deaton, and Patrick Schorderet, all postdocs in the lab of Bob Kingston.
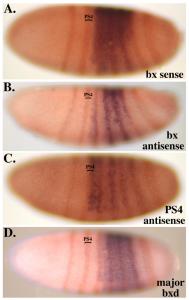
The pattern of H3K27me3 in successive parasegments reveals the segmental domains deduced from the genetic studies of Lewis. The repressed and active domains show the presence and absence of H3K27me3, respectively. The K27me3 traces from successive segments follow a “stairstep” pattern, with the adjacent active domains covering successively more of the complex in more posterior body segments (Bowman et al., 2014). The boundaries between active and repressed domains coincide with binding sites for the CTCF protein, which is proposed to be a domain boundary in many other systems. Proteins of the Polycomb Group are found at so-called “Polycomb Response Elements”, typically one per segmental domain, and these proteins are present whether the surrounding domain is active or repressed. We are extending these studies, mapping other chromosomal proteins and histone modifications, and repeating the analysis for additional segments.
Domain Boundaries
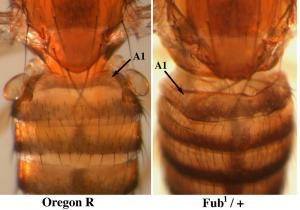
The boundaries between segmental domains are particularly interesting. They were first discovered in genetic studies; small deletions of these sites gave dominant transformations of one segment into the next more posterior segment (Bender and Lucas, 2013). Our interpretation is that the boundaries prevent the spread of the active marks of one domain into the adjacent repressed domain. These sites are nucleosome free regions, usually (but not always) coincident with CTCF binding sites. CTCF is not sufficient for boundary formation; several other proteins are involved, and they differ from one boundary to another. We are attempting to isolate nuclei from a single segment in an embryo in which a boundary has been deleted. We hope to map the proteins and histone modification associated with the spreading activation, and perhaps to watch the spreading as it happens.
Non-Coding RNAs
The 300 kb of the bithorax complex codes for only three proteins involved in segmental determination. There are, however, many non-coding RNAs, each produced in the body segments appropriate for the DNA domain from which it is transcribed. It has been argued by others that such transcripts induce the active state for the domain in which they reside. We have shown this is not the case for the best studied such transcript, made in the first abdominal segment (Pease et al., 2013). There are more subtle functions for at least one long non-coding RNA, made in the 8th abdominal segment; it encodes a micro RNA (Bender, 2008), and it represses one of the protein coding genes by transcriptional interference (Gummalla et al., 2012). We hope to test for potential functions of other non-coding RNAs by deleting or inverting their promoters.